Command Palette
Search for a command to run...
Online Tutorial: RFdiffusion2 Achieves atomic-level Protein Generation Based on Chemical Reactions, Achieving a 100% Success Rate in 41 Cases

Previously, the generative protein design model RFdiffusion primarily generated protein structures with precise fixed sites by describing ideal active sites. However, this approach had two major limitations that were difficult to overcome:
* Active site geometry can only be specified at the residue level, and researchers must determine the possible backbone positions of residues by enumerating side chain rotamers;
* The positions of catalytic residues must be predetermined in the sequence, greatly limiting the space of sampled solutions.
In response to related challenges, in April 2025, the Institute for Protein Design at the University of Washington released a new generative model, Rosetta Fold diffusion 2 (RFdiffusion2).This has achieved an optimized path for generating protein skeletons with customized active sites based on simple chemical reaction descriptions, breaking through the long-standing technical bottleneck in catalyst design.It provides strong technical support for a variety of applications including plastic degradation.
RFdiffusion2 uses a new deep generative approach that enables protein design based on sequence-independent descriptions of functional group positions, without the need for reverse rotation isomer generation. Compared to previous models, RFdiffusion2's optimization focuses on three key areas:
* Eliminated the reliance on residue enumeration and sequence indexing, and introduced flow matching and stochastic centering techniques, allowing the model to generate protein backbones directly at the atomic level;
* Supports index-free atomic-level active site generation, can process ligand information and automatically sample ligand conformations during the generation process, improving design flexibility;
* Utilizes a new AME benchmark, a computational protein design benchmark covering 41 different active sites (the previous generation only covered 16).
In further experiments, the research team designed proteins around three different catalytic sites. The experimental results showed thatRFdiffusion2 shows significant performance improvements over previous methods, successfully generating protein backbones that meet the constraints in 41 cases, with a success rate of 100%;Under the same conditions, the success rate of RFdiffusion1 was only about 39%. It can be said that the launch of RFdiffusion2 marks a key step forward in AI-driven protein design.
RFdiffusion2: A Protein Design Tool is now available on the HyperAI website (hyper.ai) in the Tutorials section. Experience the latest protein generation tool with just one click.
Tutorial Link:
https://hyper.ai/cn/tutorials/44096
Demo Run
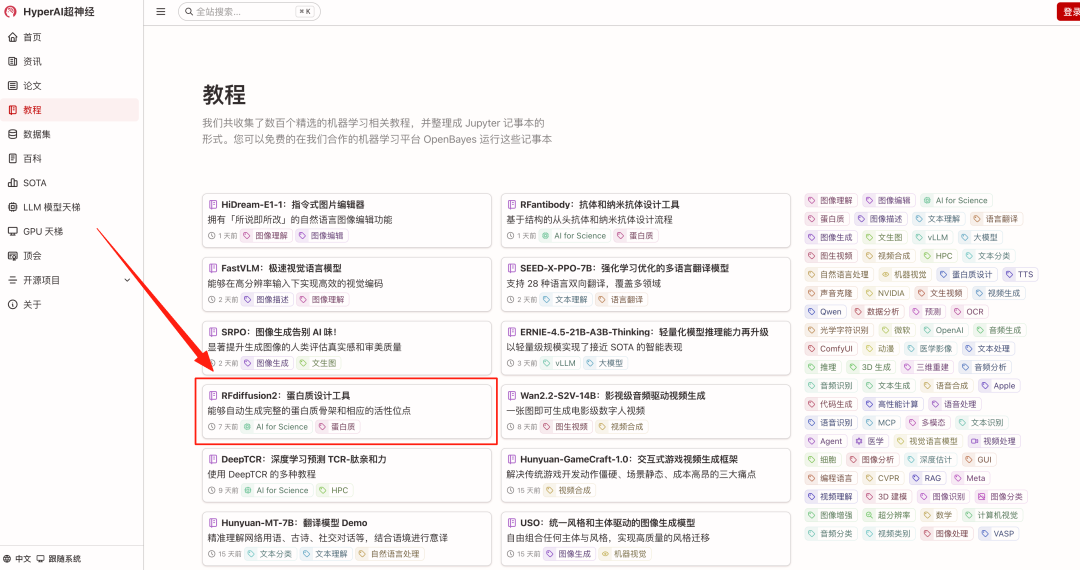
1. Enter the URL hyper.ai in your browser. Once you reach the homepage, click the Tutorials page, select RFdiffusion2: Protein Design Tool, and click Run this tutorial online.
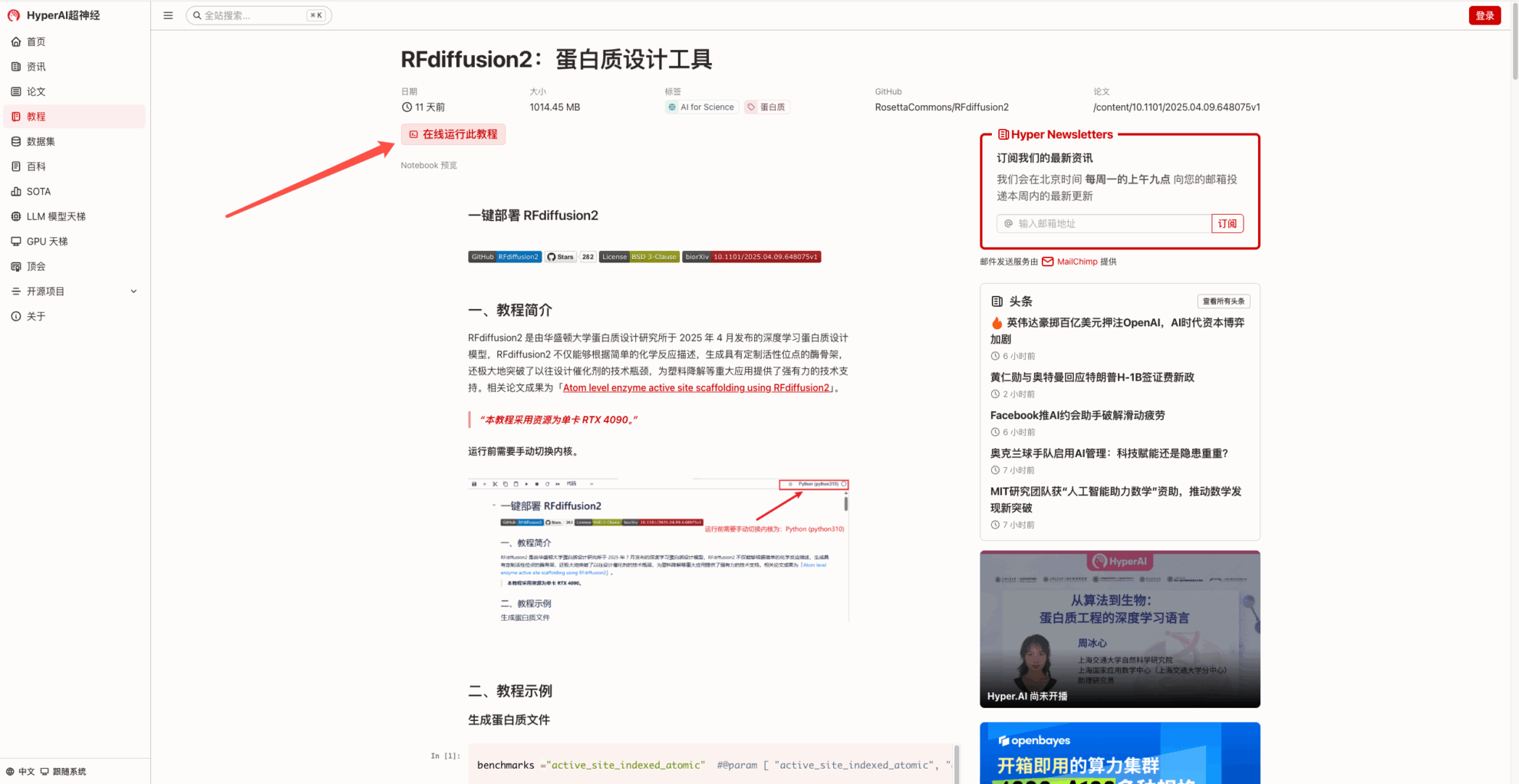
2. After the page jumps, click "Clone" in the upper right corner to clone the tutorial into your own container.
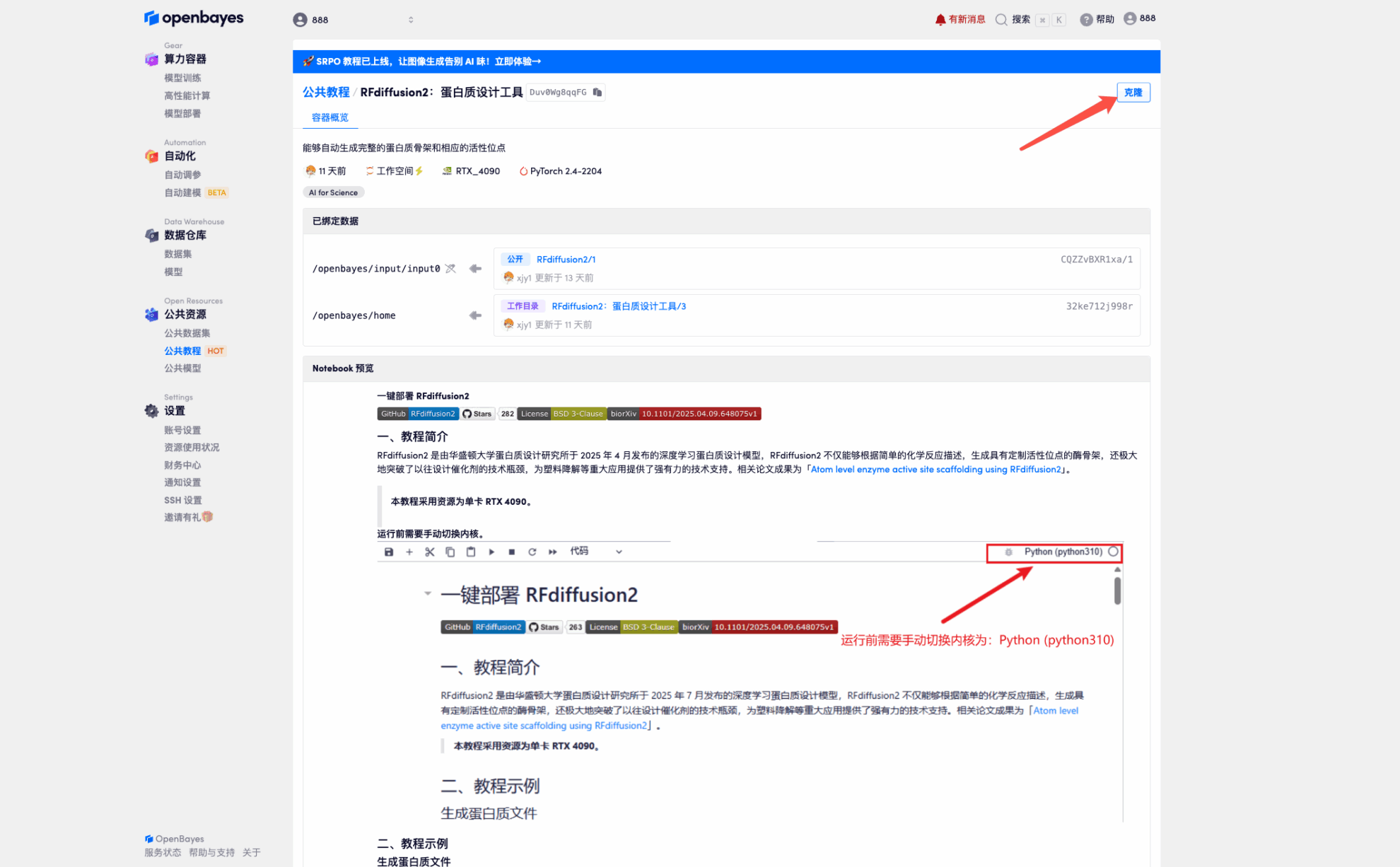
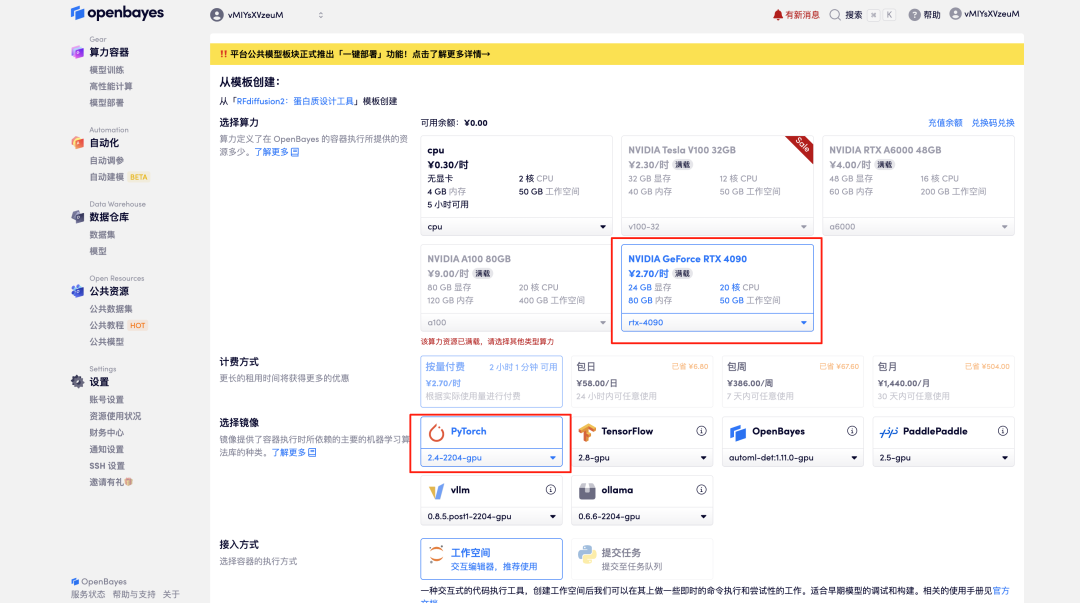
3. Select "NVIDIA GeForce RTX 4090" and "PyTorch" images, and click "Continue". The OpenBayes platform provides 4 billing methods. You can choose "pay as you go" or "daily/weekly/monthly" according to your needs. New users can register using the invitation link below to get 4 hours of RTX 4090 + 5 hours of CPU free time!
HyperAI exclusive invitation link (copy and open in browser):
https://openbayes.com/console/signup?r=Ada0322_NR0n
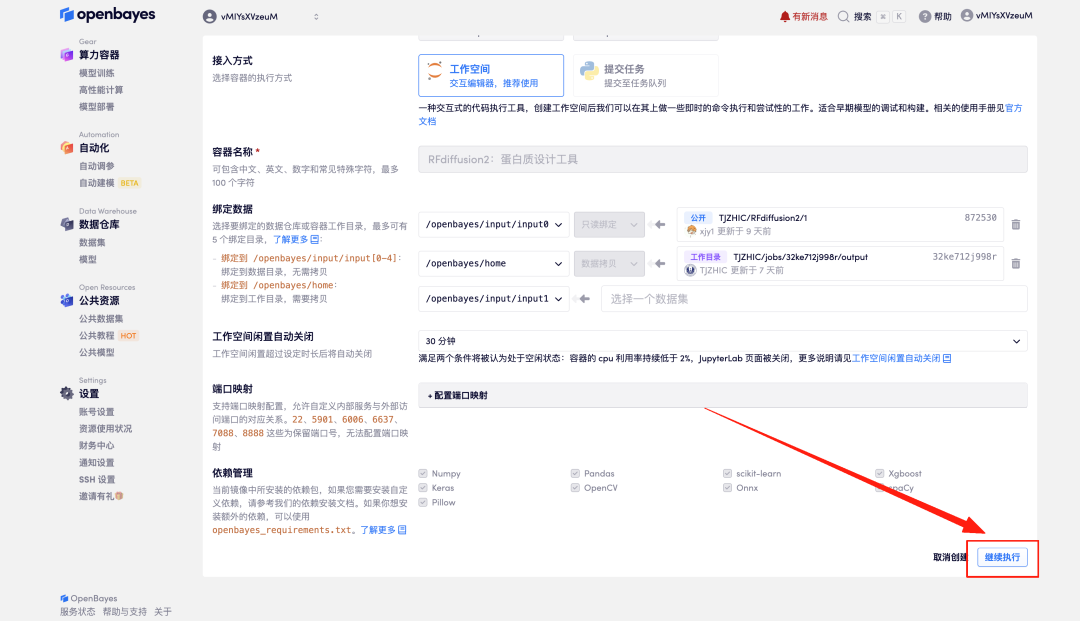
4. Wait for resources to be allocated. The first clone will take about 2 minutes. When the status changes to "Running", click the jump arrow next to "Open Workspace" to jump to the Demo page.
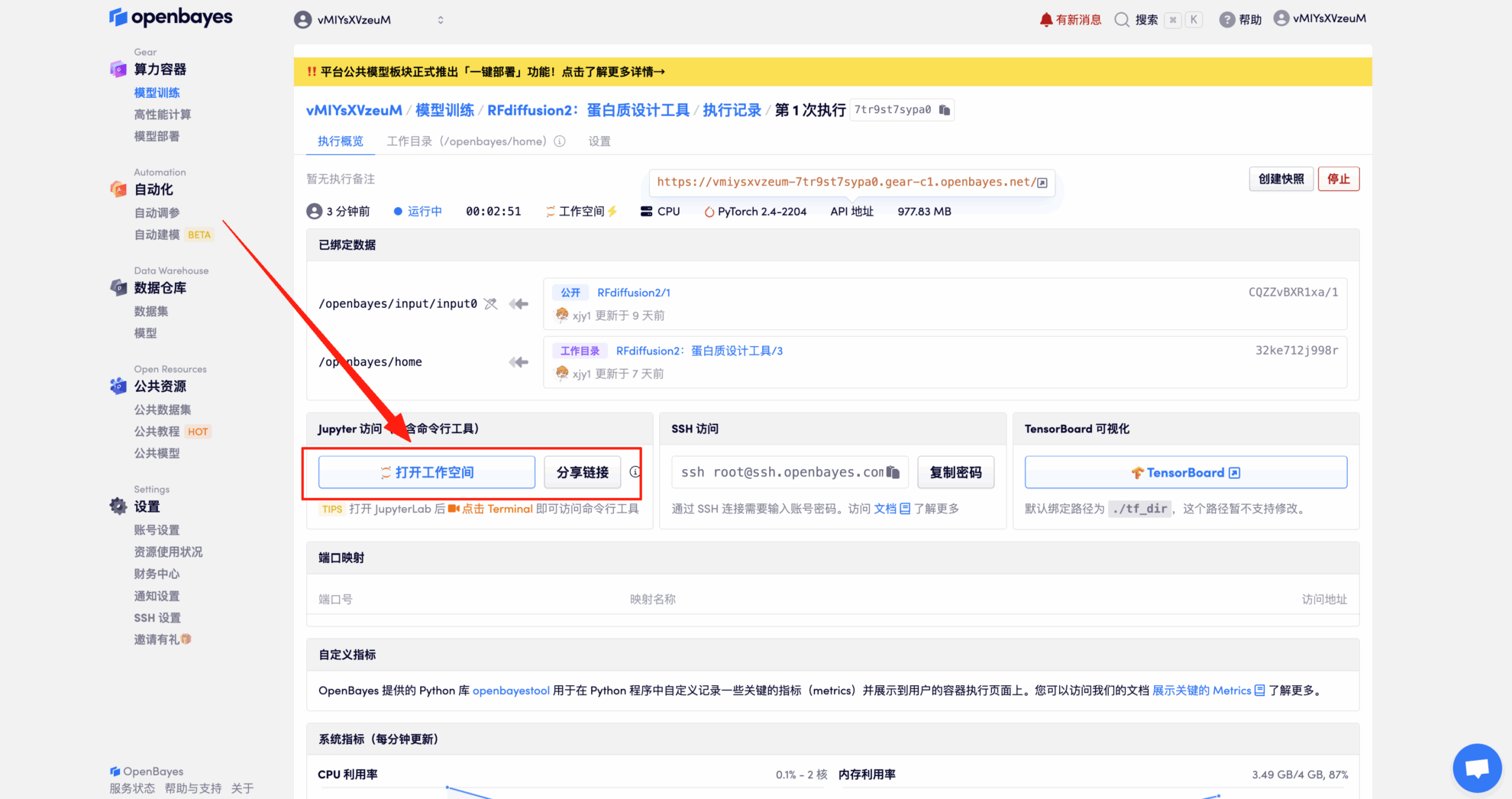
Effect Demonstration
After entering the Demo run page, enter benchmarks and click Run to get the output results.
Here we take "active_site_indexed_atomic" as an example
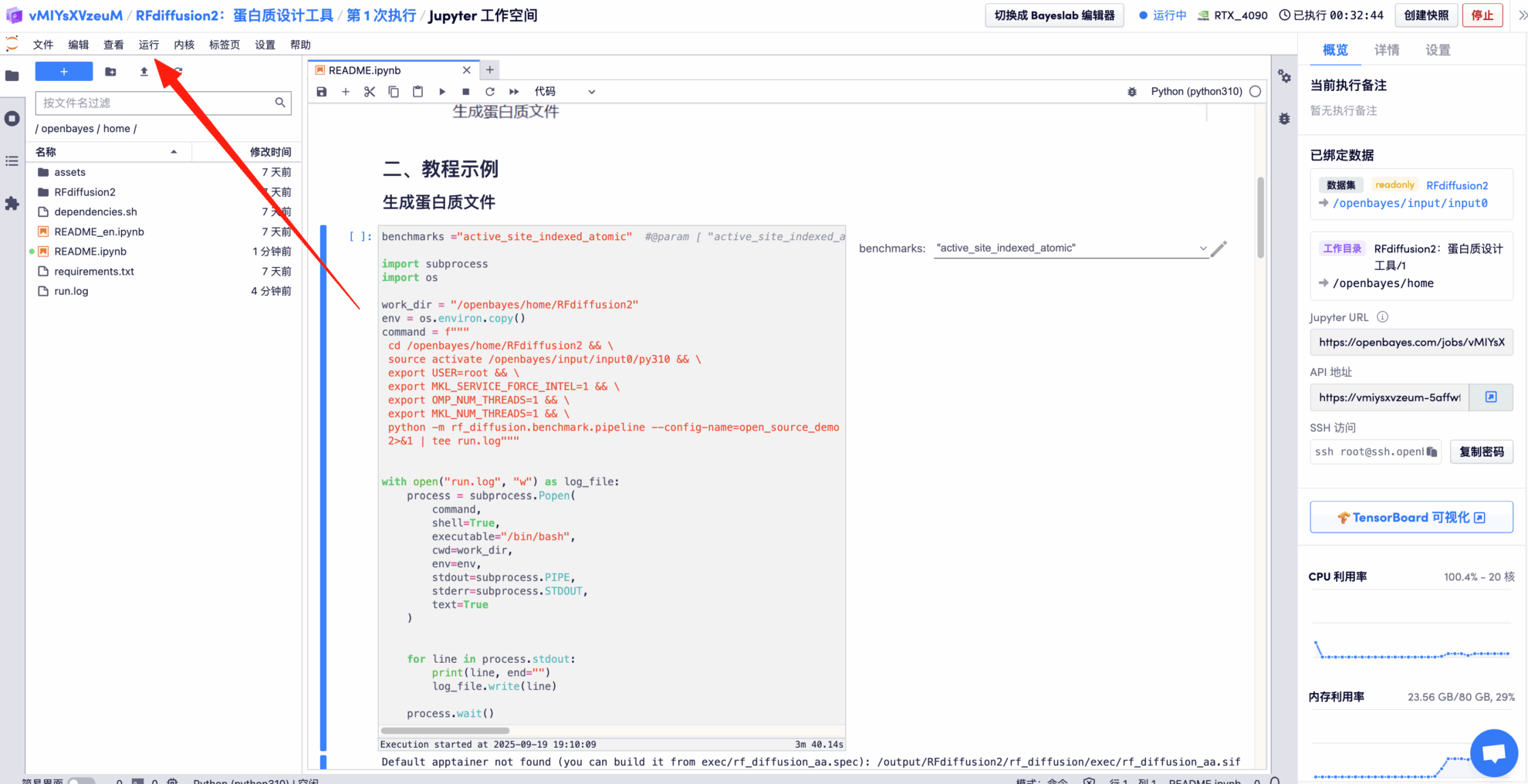
* Display the final protein design file
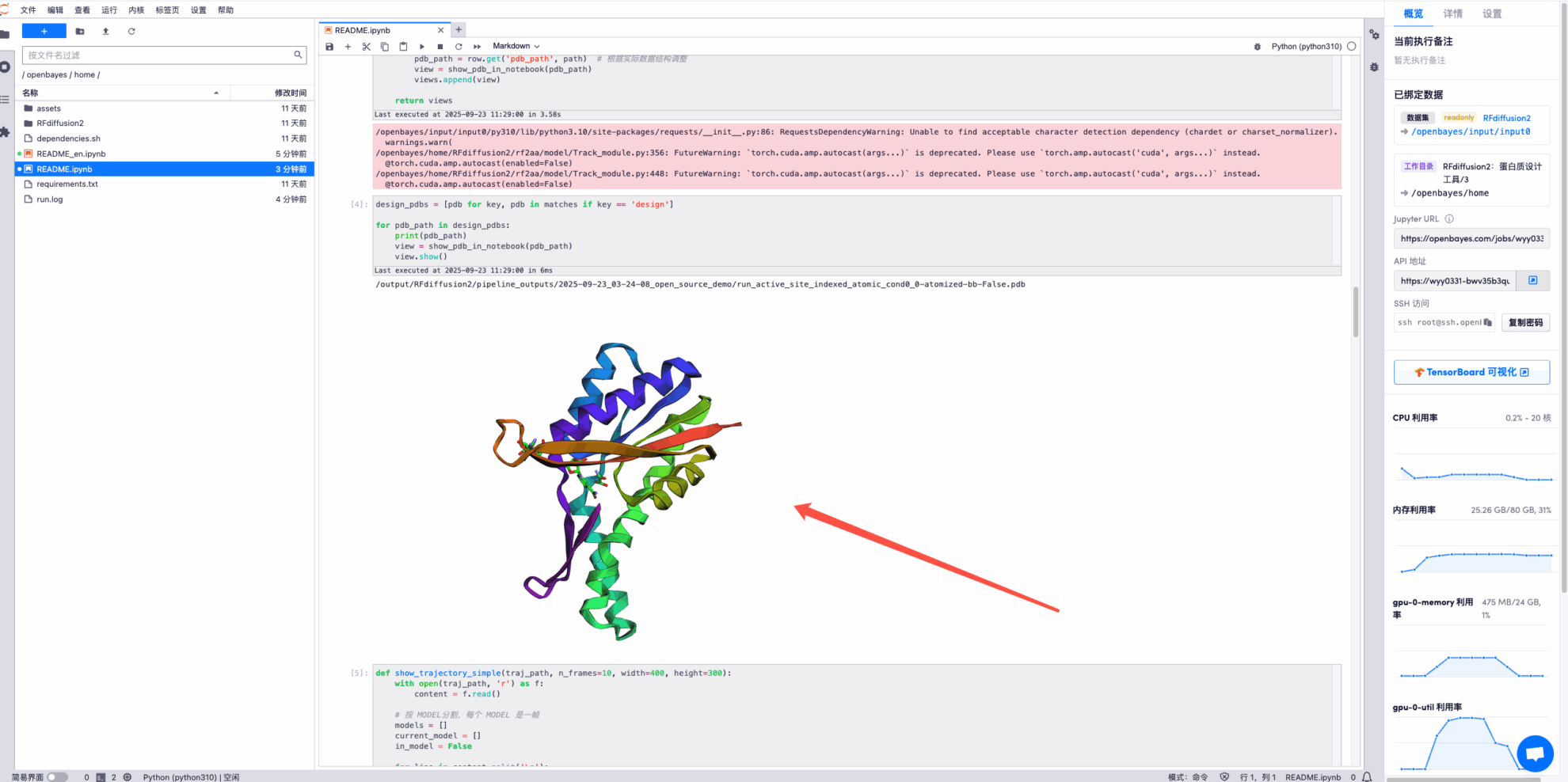
* Display the denoised structure at each time during the diffusion process. Some examples are as follows:
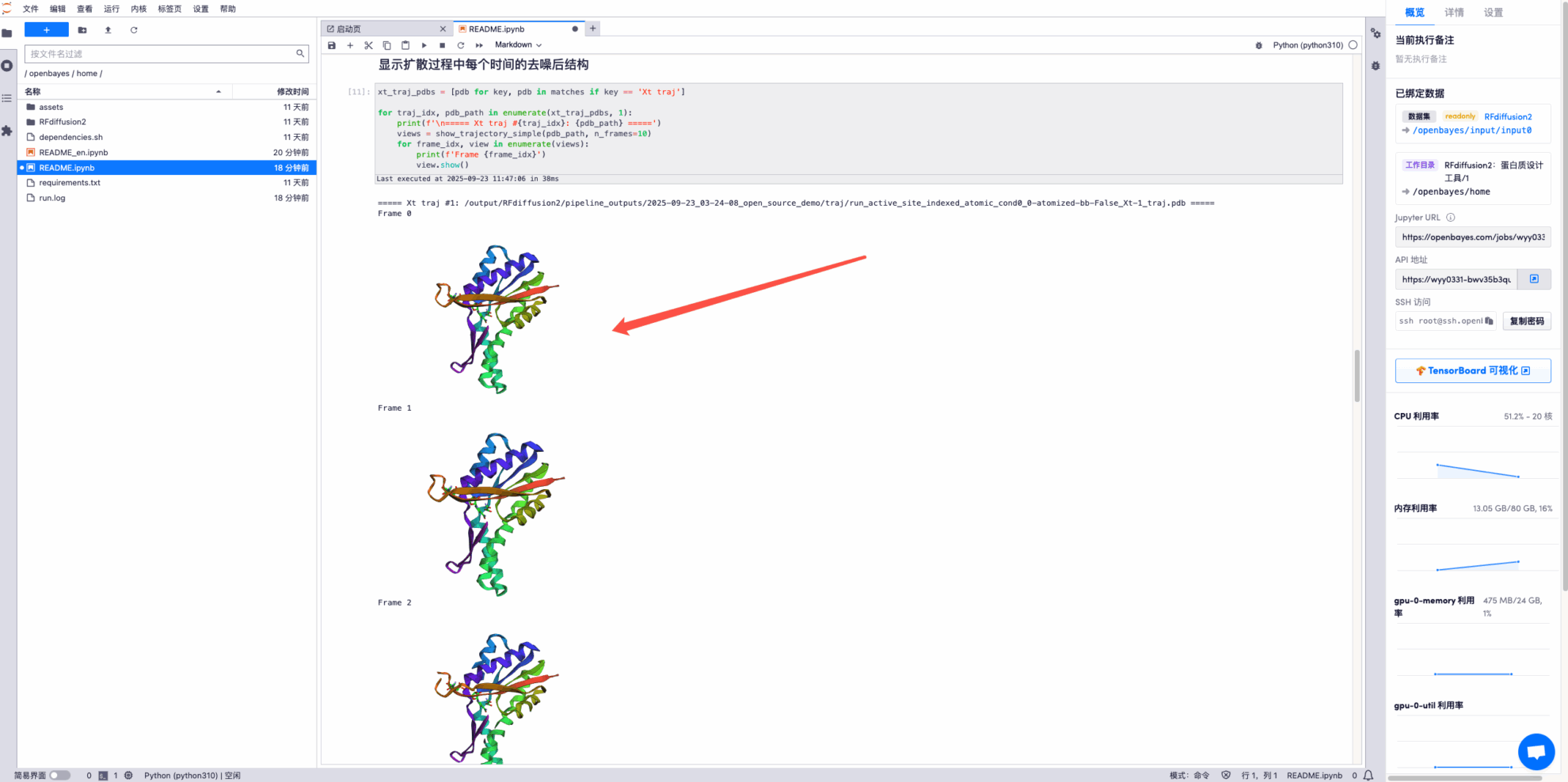
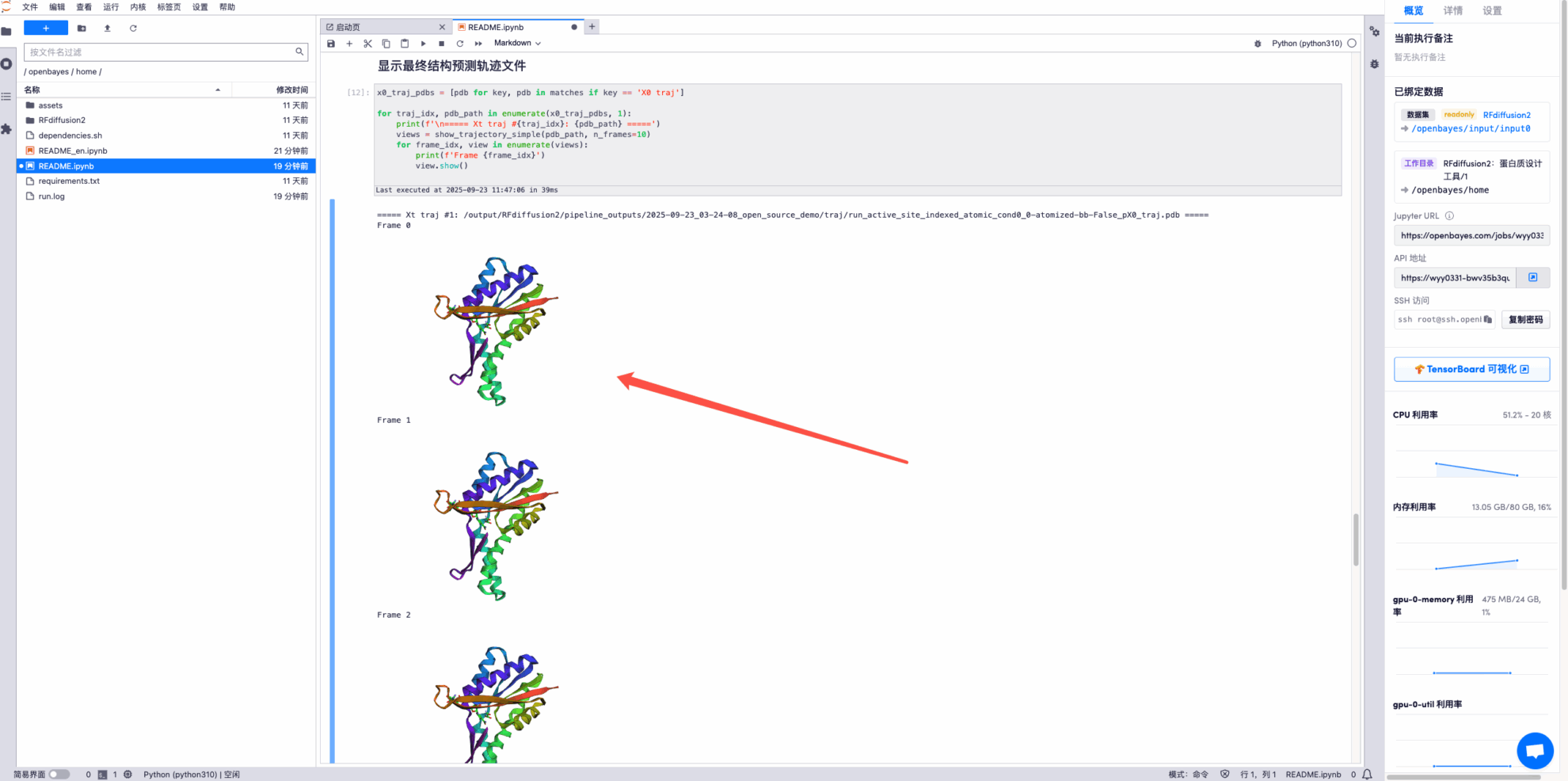
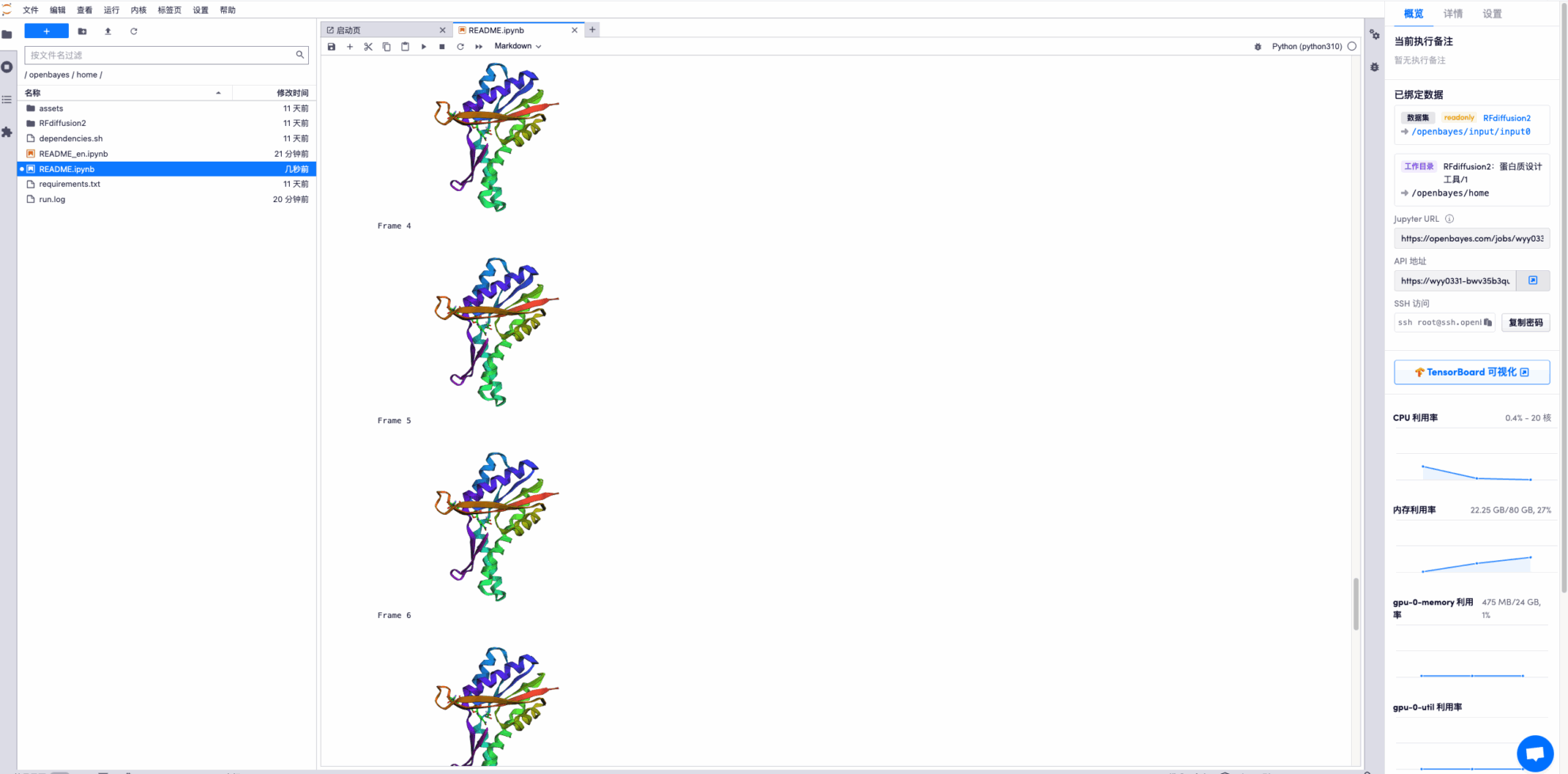
* Display the final structure prediction trajectory file, some examples are as follows:
The above is the tutorial recommended by HyperAI this time. Everyone is welcome to come and experience it!
Tutorial Link: